Postdoctoral opportunity; Translational Regulation & Innate Immunity
We have an opening for a postdoc position to investigate the mechanisms of mRNA-specific translational regulation of the innate immune system.
We’re looking for a recent PhD graduate with a strong, demonstrable background, including first-author publication(s), in the fields of RNA biology, RNA-binding proteins, and regulation of the innate immune response to viral infections. Experience with transcriptome-wide analyses of mRNA translation (e.g. Ribo-Seq) or RNA-protein interactions (e.g. CLIP) would be a bonus.
Apply via this link: https://hrwebapp.qub.ac.uk/tlive_webrecruitment/wrd/run/ETREC107GF.open?VACANCY_ID=004407UfFV&WVID=6273090Lgx&LANG=USA
For further information on the research background, see the following references:
1. Choi JH, et al. Repression of mRNA translation initiation by GIGYF1 via disrupting the eIF3-eIF4G1 interaction. Sci Adv. 2024 Jul 19;10(29):eadl5638.
2. Zhang X, et al. SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation. PNAS. 2022 Aug 9;119(32):e2204539119.
3. Zhang X, et al. microRNA-mediated translational control of antiviral immunity by the cap-binding protein 4EHP. Molecular Cell 2021 Feb 9:S1097-2765(21)00050-2.
4. Álvarez L, et al. The molecular dissection of TRIM25’s RNA-binding mechanism provides key insights into its antiviral activity. Nat Commun. 2024 Oct 1;15(1):8485.
5. Kamel W, et al. Alphavirus infection triggers selective cytoplasmic translocation of nuclear RBPs with moonlighting antiviral roles. Mol Cell. 2024 Dec 19;84(24):4896-4911.e7.
Congratulations to Susanta for his Cancer Research UK Bridge to Academic Leadership Award
We are delighted to congratulate Susanta on being awarded the highly competitive Cancer Research UK Bridge to Academic Leadership Award. This award recognises Susanta’s outstanding research, independence, and potential as a future leader in cancer research. It provides a platform to support his transition towards an independent academic career, and we are incredibly proud to see his hard work and dedication recognised in this way.

A Fond Farewell to Patric
We wish a fond farewell to Patric, who has recently moved on to begin his postdoctoral training at the University of Oxford. During his time in the lab, he was an outstanding student, thoughtful, and always generous with his time and ideas. He made a real impact on both our research and the lab environment, and he will be greatly missed by everyone here. We are incredibly proud of all he has achieved and wish him every success in this exciting next stage of his career.

Competitive PhD Studentship open to both domestic and international students.
We are inviting applications for a fully funded 4-year PhD studentship through the BBSRC-funded NorthWestBio PhD Programme. The project is a collaboration with Alfredo Castello‘s group at University of Glasgow. The successful candidate will be primarily based in Glasgow, with opportunities to interact closely with collaborators in Belfast.
This multidisciplinary project sits at the intersection of RNA biology and the host-pathogen response, focusing on the mechanisms of translational regulation mediated by GIGYF1/2 proteins in controlling host responses to RNA virus infections.
The successful candidate will lead a cutting-edge research project in RNA biology and virology in a highly collaborative environment across two leading research groups. You will receive training in molecular biology, virology, translational regulation, and transcriptome-wide analysis of mRNA translation and RNA-protein interactions.
For further information on the research background, see the following references:
1. Álvarez L, et al. The molecular dissection of TRIM25’s RNA-binding mechanism provides key insights into its antiviral activity. Nat Commun. 2024 Oct 1;15(1):8485.
2. Kamel W, et al. Alphavirus infection triggers selective cytoplasmic translocation of nuclear RBPs with moonlighting antiviral roles. Mol Cell. 2024 Dec 19;84(24):4896-4911.e7.
3. Iselin L, et al. RNA binding regulation is a new dimension in the type I IFN response. bioRxiv, 2025.
4. Choi JH, et al. Repression of mRNA translation initiation by GIGYF1 via disrupting the eIF3-eIF4G1 interaction. Sci Adv. 2024 Jul 19;10(29):eadl5638.
5. Zhang X, et al. SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation. PNAS. 2022 Aug 9;119(32):e2204539119.
6. Zhang X, et al. microRNA-mediated translational control of antiviral immunity by the cap-binding protein 4EHP. Molecular Cell 2021 Feb 9:S1097-2765(21)00050-2.
Submit your application directly through the NorthWestBio portal by November 21st. Shortlisted candidates will be invited for interview.
The QUB contingent, including members of the RNA Biology group attending the 3rd Irish RNA Club annual meeting at Maynooth University.

PhD studentship
We are excited to offer a competitive PhD studentship in my group and in collaboration with Colin Adrain and Graeme Greenfield.
The studentship is funded by Leukaemia & Lymphoma NI (LLNI), who will pay stipend and tuition fees only at the UK/ROI level. However, there are no restrictions on the nationality of the LLNI funded PhD student.
Find more info about how to apply here.
This project will investigate how mRNA translation and protein synthesis quality control mechanisms influence AML cell survival and activation of the immune response under chemotherapy-induced stress. Using advanced molecular biology techniques—including CRISPR genome editing, high-throughput analysis of mRNA expression and translation (RNA-Seq and Ribo-Seq), and proteomics—you will identify key regulatory proteins in cellular quality control pathways that protect AML cells, then determine whether targeting them can enhance chemotherapy efficacy. Additionally, you will explore how disrupting these pathways influences the ability of AML cells to be “visible” to the immune system, potentially leading to new immunotherapeutic strategies.
Don’t hesitate to get in touch and also check some of our recent publications (see links below) for more info.
https://academic.oup.com/nar/article/52/20/12534/7798793
https://www.pnas.org/doi/abs/10.1073/pnas.2413018121
https://www.science.org/doi/full/10.1126/sciadv.adl5638
https://febs.onlinelibrary.wiley.com/doi/full/10.1111/febs.17217
New Preprint
We’re excited to share our latest study, now available on bioRxiv, uncovering a new mechanism in higher eukaryotes that connects the dots between codon usage, translation efficiency, and mRNA stability.
In this work, we investigated how translation of non-optimal codons—especially those ending in A or U at the wobble position (A/U3 codons)—can impact the stability of mRNA. While it’s known that slower translation elongation at non-optimal codons can lead to mRNA degradation, the molecular players that sense and act on this in higher eukaryotes (e.g. mammals) have remained elusive.
Our study identifies two paralogous RNA-binding proteins, ZC3H7A and ZC3H7B, as key regulators in this process. Found only in chordates, these proteins preferentially bind to mRNAs rich in A/U3 codons, where they either trigger degradation or block translation initiation. This is achieved through two distinct interactions:
- With the CCR4-NOT complex, promoting mRNA decay
- With the GIGYF2/4EHP complex, repressing translation initiation
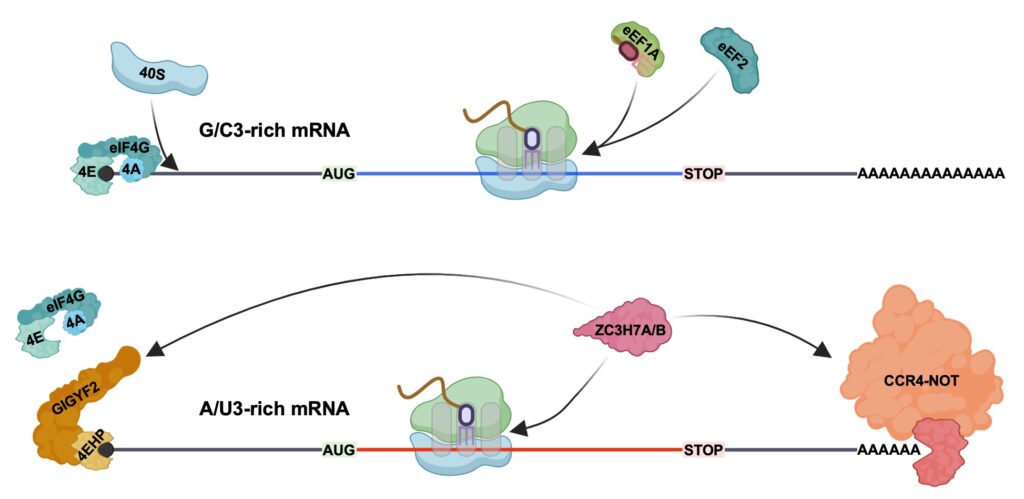
These findings reveal a dual mechanism through which ZC3H7A/B tune gene expression based on codon usage—a remarkable example of how organisms have evolved to fine-tune protein synthesis beyond the genetic code itself.
🎉 A huge congratulations to Patric and Parisa for their outstanding work driving this study forward. And many thanks to all our collaborators, particularly our bioinformatics guru Sarah Maguire, for their invaluable contributions—this was truly a team effort!
Stay tuned for more as we continue to explore the layers of post-transcriptional regulation that shape gene expression.
Apply for a The Newton International Fellowships
The Newton International Fellowships for this year are now open for applications! If you’re a non-UK scientist looking to pursue a postdoc in the UK, you’ll need a host lab. If you’re interested in RNA biology, feel free to reach out to discuss the possibility of joining my lab. Check the link below for more details, including eligibility criteria, deadlines, etc. https://royalsociety.org/grants/newton-international/












